Unlocking the power of data for scientific discovery
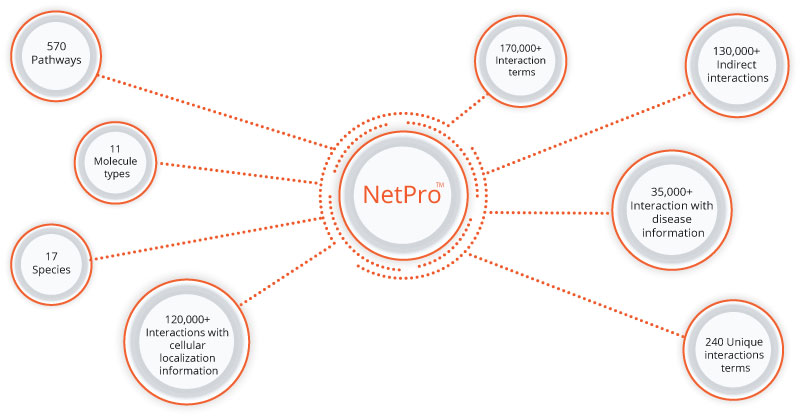
NetProTM currently has more than 320,000+ manually curated unique interactions from scientific literature. The knowledgebase facilitates navigation through the complex human biological pathways map including metabolic processes and cellular signaling.
The strength of NetProTM lies in the manual curation of the interactions and in the content granularity. The knowledgebase has been extensively normalized with standard terms for both the interacting molecules and the type of interactions.
Exhaustive controlled vocabulary largely sourced from public domain makes it easier for the knowledgebase to be integrated to other proprietary databases.
Molecule Type (Protein, DNA, RNA, etc.)
Domain information on proteins
Species information
Interaction types
Cellular localization
Experimental methods used to determine the interaction
Mediators and regulators of the interaction
Pathway information
Kinetic information
Diseases related information
- Covers several entities other than proteins as interacting partners, like RNA, DNA, etc. with well defined, exhaustive interaction terms
- Resource for target identification, validation and disease specific pathway building
- Quantitative/Qualitative alterations in an interaction due to a pathological condition(s), polymorphism, mutation, post Translational Modifications or other experimental conditions
- Comprehensive tool identify upstream, downstream mediators and regulators of the interaction
Protease module
Kinase module
Transcription factor module
Bacterial/Viral interaction module
Phosphatase module
Small molecule/Drug module
Ubiquitination module
Cytochrome P450 module
- The knowledgebase can be specifically queried and filtered for various fields like molecules, molecule types, species, interaction terms, interaction type, cellular localization, pathway, disease etc.
- Disease related information pertaining to the interactions is available which could be a mutation detail, expression profile or any other significant properties of the interacting molecule with respect to the disease.
- MINETM a Java based visualization tool developed by Molecular Connections, could be used for querying, filtering and analyzing NetProTM data.
- Molecule IDs are mapped to a number of external identifiers like UniProt, Gene RefSeq etc. in case of proteins and Pfam IDs incase of domains, PubChem in the case of small molecules.
- The gene IDs are mapped to their corresponding probe IDs, making the analysis of micro array data possible.
- The disease terms used in NetProTM are normalized using MeSH ontology while the cell lines are ATCC normalized. Pathway captured are also GO compliant.
- Extensive normalization makes NetProTM user friendly with respect to data integration into internal proprietary databases.
Dive Deeper
Request additional information on NetPro™
Got questions? Need more info? Fill out the form and our team will get back to you soon.
